Current projects
Computational analysis of the relationship between histone modifications and gene expression
Rosa Karlić, Kristian Vlahoviček
This project studies histone modifications, or modifications of proteins which participate in packing of DNA and are closely linked to gene regulation. There is evidence that histone modifications are in fact a mean for coding for certain processes downstream of genes. Quantative models for predictions of gene expression levels through histone modification have shown thus far that gene expression and histone modification have a high correlation and that a relatively small number of histone modifications are necessary to predict gene expression levels, as well as that their relationship is general and valid in many cell types.
A mutational signature reveals alterations underlying deficient homologous recombination repair in breast cancer.
Analysis of somatic microsatellite indels identifies driver events in human tumors. Nature biotechnology, 2017
Cell-of-origin chromatin organization shapes the mutational landscape of cancer. Nature, 2015
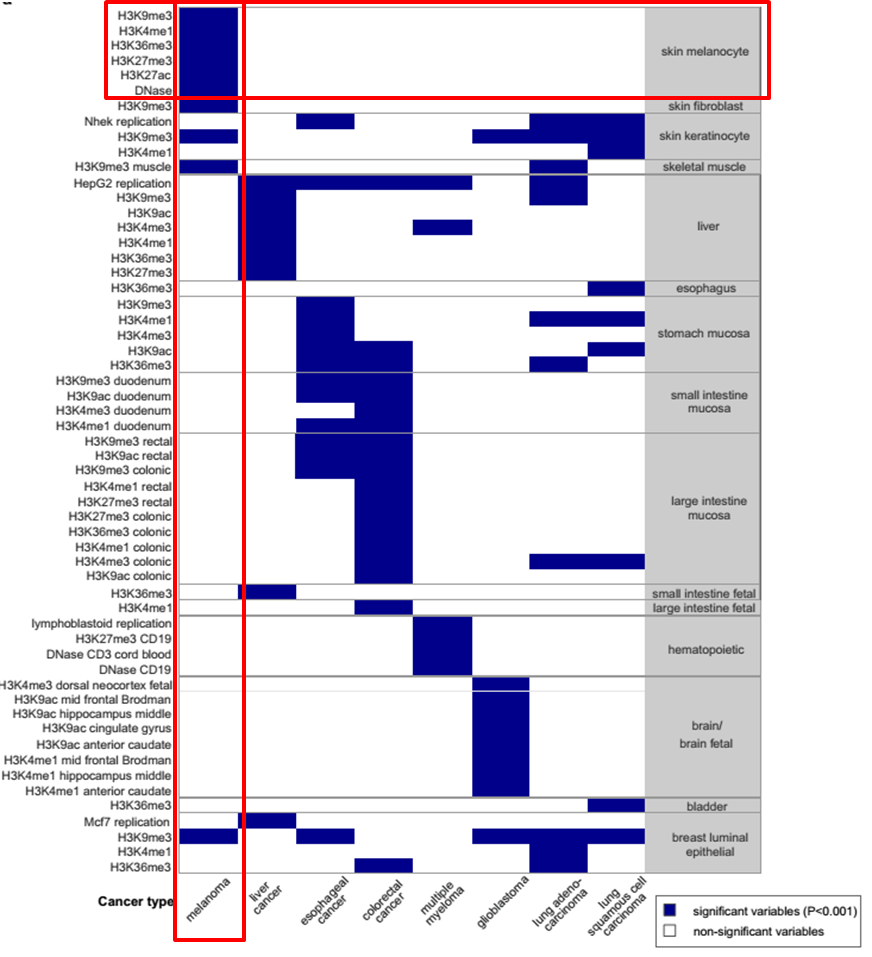
Integrative Analysis of 111 Reference Human Epigenomes. Nature, 2015
Histone modification levels are predictive for gene expression. PNAS,
2010
Genomic complexity of the simplest Metazoa, the sponge
Zoe Jelić Matošević, Maja Kuzman, Maša Roller, Kristian Vlahoviček
Sponges are living fossils, inhabiting the planet for 580 million years, and the closest living species to the first multicellular animal. Therefore, by researching sponges we are also investigating our own evolutionary past. Our research of sponge genomes has shown that, despite the morphological simplicity of sponges, their genetic repertoire is surprisingly complex. This shows that even ancient ancestors of animals already had the potential for more complex life functions. The focus in evolutionary studies is shifting from the increase of genome complexity to adaptations which enable exploitation of the existing complexity.
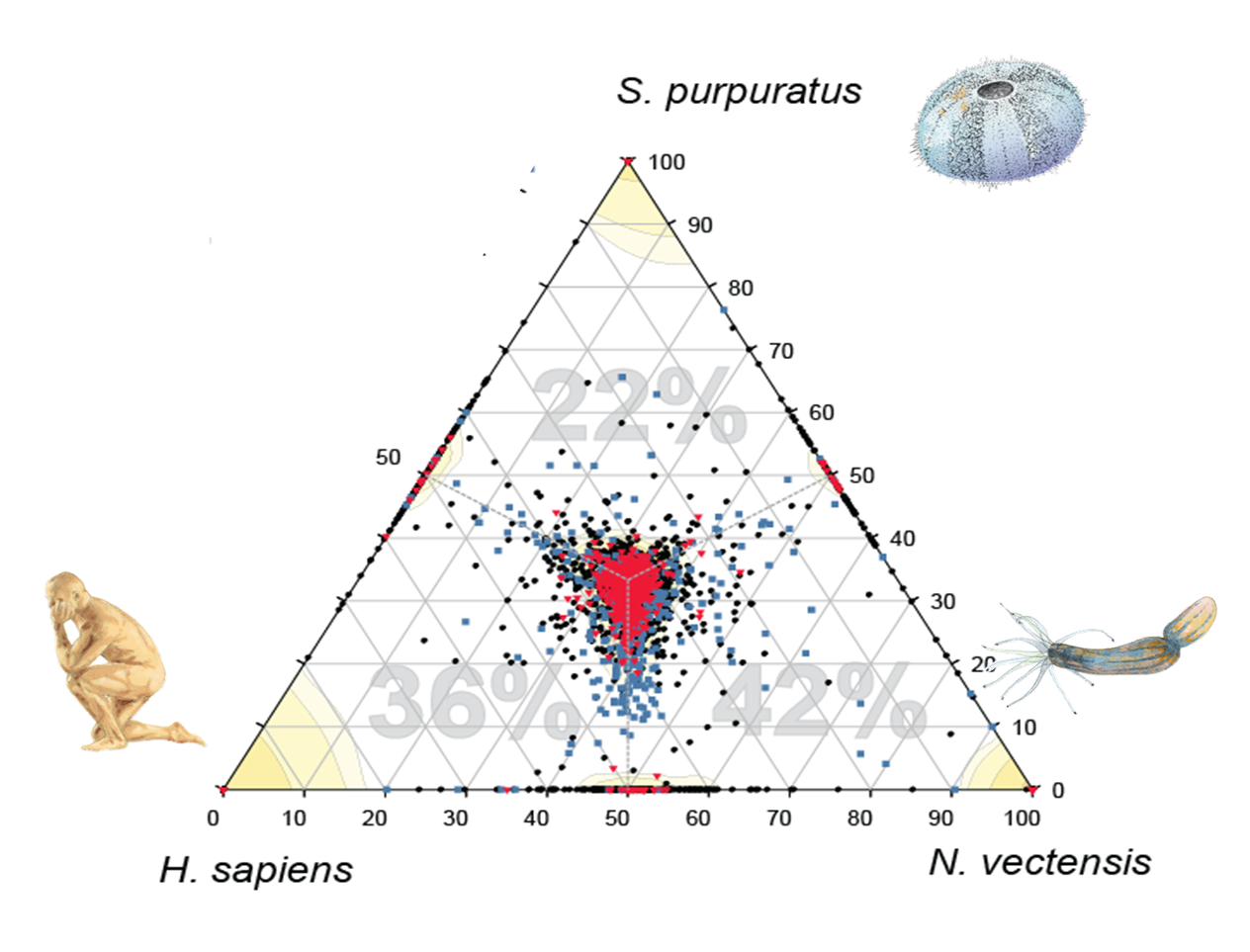
Demosponge EST sequencing reveals a complex genetic toolkit of the simplest metazoans.
Over-represented localized sequence motifs in ribosomal protein gene promoters of basal metazoans.
Codon usage in metagenomes
Maja Kuzman, Vedran Lucić, Maša Roller, Kristian Vlahoviček
Metagenomics, the study of bacterial communities isolated straight from the environment, is a field which investigates bacterial populations in their natural environments. We identify functional adaptations of whole bacterial communities which enable them to survive in diverse environmental conditions such as animal digestive tracts, where bacteria degrade food into forms which can be harvested by the animal and where changes in bacterial composition can lead to obesity and disease. The method we are developing can not only identify the functions bacteria posses but also predict how much they are used and therefore offer a more accurate description of life in bacterial communities.
Big Data, Evolution, and Metagenomes: Predicting Disease from Gut Microbiota Codon Usage Profiles.
Environmental shaping of codon usage and functional adaptation across microbial communities.
Translational Selection Is Ubiquitous in Prokaryotes.
Comparison of codon usage measures and their applicability in prediction of microbial gene expressivity.
