Teaching activities
Teaching activities are held in form of graduate courses as well as annual courses in collaboration with some European experts. Our teaching is compliant with the guidelines of Bologna convention and we extensively use distance-learning methods. We have one introductory course – Bioinformatics, while the rest of the courses are a part of Computational biology module for studding advanced bioinformatics tools and methods.
Communication with students and distribution of course materials are accomplished through our bioinformatics eduactional portal, BEPo. The portal is based on the “Moodle” e-learning system and has been in use for over a decade.
Algorithms and programming
Introduction to R
R, R markdown, R notebook Vectors, Matrices, Functions Flow control in R Lists Factors
Data manipulation
Data frames Package: dplyr Package: data.table Regular expressions Package: tidyr Cleaning your data
Data visualization
Some useful graphs: Scatter plot Q-Q plot Histograms Density plots correlation matrix (package:corrplot) Heatmap (package: pheatmap) Package: ggplot2 Interactive graphs
Advanced topics: Bioconductor
Package:Biostrings
Package:shortRead
Package: biomaRt
Package: GenomicRanges and IRanges
Visualization of genomics data: ggbio, sushi, circos
karyograms
circular plots
alignments
Literature:
Hadley Wickham (2014) Advanced R
R. Kabacoff (2011) R in Action 1st ed. Manning Publications
Machine learning and statistics
Lectures
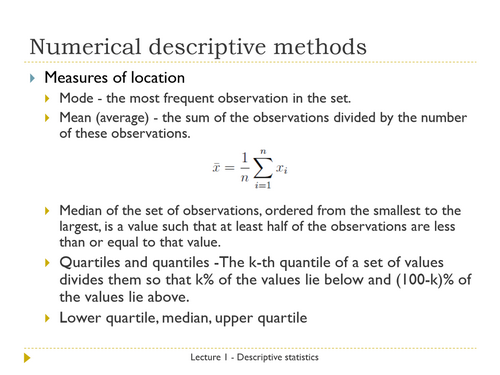
1. Introduction to data, experimental design and descriptive statistics: populations and samples, measurement scales, categorical and numerical variables, descriptive statistics, numerical analysis of distributions – measures of location and spread, graphical analysis of distributions – pie-charts, bar plots, histograms, box and whisker plots.
2. Confidence intervals and hypothesis testing: p-values, Student’s t-test, nonparametric statistical tests, contingency table analysis.
3. Statistical tests: Chi-square test, Fisher’s exact test, hypergeometric test.
4. Statistical power and multiple hypothesis testing: type I (α) and type II (β) errors.
5. Analysis of variance (ANOVA).
6. Correlation and linear regression: Simple and multiple linear regression.
7. Linear models for classification.
8. Model assessment and selection – resampling methods (bootstrap, cross-validation), feature selection methods, regularization methods (ridge regression, LASSO, elastic net).
9. Support vector machines.
10. Tree-based methods: regression and classification trees, bagging, boosting, Random Forests.
11. Clustering methods: hierarchical clustering, k-means clustering.
12. Dimensionality reduction methods: Principal components analysis (PCA).
Practical
1. Analysing distributions in the R statistical environment. Calculating the measures of location and spread implemented in the base package.
2. Analysing distributions in the R statistical environment. Functions for graphical analysis of samples.
3. Calculating confidence intervals. Hypothesis testing and errors in hypothesis testing (Type I and Type II errors). Power of a statistical test.
4. Comparison of different types of Student’s t-test (based on the dependence of the observations in different samples, the size of the samples and the equality of variance of the samples) in the R statistical environment.
5. Implementation of Fisher’s test, chi-squared test and hypergeometric test in the R statistical environment.
6. Analysis of linear regression models. Interactions. Qualitative predictors. Transformation of non-linear regression models. Analysis of residuals.
7. Implementation of analysis of variance in the R statistical environment.
8. Comparison of various linear models for classification (logistic regression, linear discriminant analysis, quadratic discriminant analysis) in the R statistical environment.
9. Comparison of ridge regression, LASSO and elastic net in the R statistical environment (glmnet package). Choosing the optimal value of λ.
10. Implementation of resampling methods in the R statistical environment (k-fold cross-validation, leave-one-out cross-validation).
11. Tree-based methods in the R statistical environment (tree, gbm and randomForest packages).
12. Support vector machines in the R statistical environment (e1071 package). Using cross-validation to select the optimal model parameters.
13. Using Hidden Markov Models to analyse biological sequences in the R statistical environment.
14. Principal components analysis in the R statistical environment (prcomp() function).
15. Clustering methods in the R statistical environment (hclust() and kmeans() functions). Choosing the optimal number of clusters.
Literature
• G. James, D. Witten, T. Hastie and R. Tibshirani , “An Introduction to Statistical Learning, with applications in R” (Springer, 2013)
• D. Montgomery and G.Runger.Applied statistics and probability for engineers (John Wiley & Sons, Inc., 2003)
Computational genomics
Lectures
1. Introduction to computational genomics: the history of (human) genome, next generation sequencing methods: Roche 454/pyrosequencing, Illumina GAII, Illumina Hiseq, ABI Solid, Helicos, Pacific bioscience, advantages and disadvantages of various sequencing platforms, file formats (GFF/GTF, BED).
2. Experimental design and quality control of sequencing experiments: controls, replication, randomization, blocking, barcoding, pre-processing of short reads obtained using next-generation sequencing, adapter removal, filtering, FastQC, samtools.
3. Genome assembly: De novo genome assembly methods and algorithms, greedy algorithms, overlap layout consensus methods, de Brujin graphs.
4. Read mapping: mapping short reads to previously assembled genomes (hashing, seed and extend, spaced seeds, algorithms based on preffix/suffix tries, Burrows Wheeler transform), Bowtie, BLASR, minimap, SAM/BAM file formats.
5. Methods for transcriptome analysis based on next-generation sequencing (RNA-Seq).
6. Methods for analyzing interactions of proteins and DNA (chromatin immunoprecipitation combined with next-generation sequencing, ChIP -Seq).
7. DNA methylation and 3D chromatin structure: Determining the three-dimensional structure of the genome (3C, 4C, 5C and Hi-C methods).
Practical
1. Introduction to genome browsers (UCSC Genome Browser).
2. Introduction to Bioconductor, a repository of R packages used for analysing biological data.
3. Manipulating and analysing strings with the Biostrings package.
4. Pattern matching.
5. – 6. Interval operations. IRanges and GenomicRanges packages.
7. – 8. Retrieving annotations from publicly available databases. GenomicFeatures package.
9. – 10. Reading and analysing short reads obtained using next generation sequencing methods. ShortRead and RSamtools packages.
11. Filtering and quality control of next generation sequencing data.
12. Normalization of next generation sequencing data.
13. Analysis of differentially expressed genes. DESeq, EdgeR and DEXSeq packages.
14. Gene set enrichment analysis and Gene Ontology enrichment analysis.
15. Methods for analysing ChIP-Seq data. Determining the genomic location of protein binding sites.
Literature
• R. C. Deonier, S. Tavare, M. S. Waterman, Computational Genome Analysis: An Introduction. Springer 2005.
• R. Gentleman, V. Carey, W. Huber, R. Irizarry, S. Dudoit, Bioinformatics and Computational Biology Solutions Using R and Bioconductor. Springer 2006.
• N. Cristianini, M. Hahn, Introduction to Computational Genomics: A Case Studies Approach. Cambridge University Press, 2007.
Bioinformatics
Lectures
1. Introduction
2. Biological databases, network resources and using specific databases.
3. Basic sequence alignment tools, substitution matrices and dynamic programing.
4. Dotplots.
5. Search by sequence similarity, Heuristic methods.
6. Tools and methods for multiple sequence alignment.
7. Patterns and motif search.
8. Basic phylogenetics.
9. Structure databases and modeling, visualization of biological macromolecules.
10. Basics of functional genomics, DNA Sequencing, Genome browsers.
Practical
Students work in groups and with assistance from professor and assistants perform research on specific topics.
While going through their projects, students search biological databases to investigate epidemiology, genetics and molecular mechanisms of he disease. After literature search, students search sequence databases and learn to manipulate with sequence information using different bioinformatics tools to acquire further information. Students complete their projects by preforming phylogenetic analysis and calcification and visually presenting their results.
Literature
• A.M. Campbell, L.J. Heyer (2002) Discovering Genomics, Proteomics and Bioinformatics. J.H.Wiley & Sons
• N. C. Jones, P. A. Pevzner (2004) An Introduction to Bioinformatics Algorithms. MIT Press
• D.W. Mount (2004) Bioinformatics: Sequence and Genome Analysis 2ed. CSHL Press
• S. B. Primrose, R.M. Twyman (2003) Principles of Genome Analysis and Genomics 3ed. Blackwell Publishing
• R. Durbin, S. Eddy, A. Krogh, G. Mitchinson (1998) Biological Sequence Analysis. Cambridge University Press.
• P. Baldi, S. Brunak (2002) Bioinformatics: A Machine Learning Approach. MIT Press